Current Projects
AI-based identification of cellular phenotypes in genetic screens
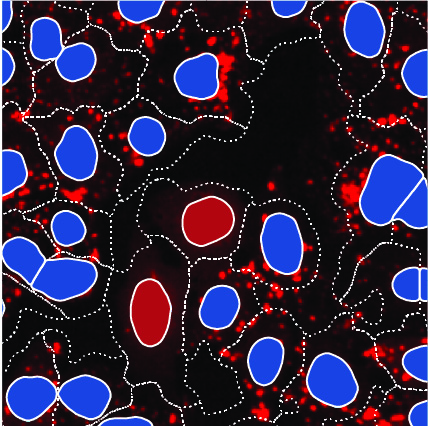
Using my SPARCS technology, I conduct image-based genetic screens in cellular model systems at human genome scale. These screens create datasets of hundreds of millions of single-cell fluorescence images. Identifying cells with relevant phenotypes in these datasets is a challenge.
I use self-supervised learning to train AI models to identify interesting phenotypes based on single-cell images. These models learn to identify specific genetic effects, enabling me to discover new gene functions.
Modeling cellular behaviour across data modalities with scPortrait
Comprehensive computational modeling of a cell's responses to its surroundings — often called "virtual cell" — requires integrating different data modalities into a single model. One modality for which data can be acquired at a scale compatible with modern AI techniques is microscopy images.
Working with Sophia Mädler and the labs of Fabian Theis and Matthias Mann labs, I developed scPortrait, a software package that processes raw microscopy fields-of-view into standardized single-cell image datasets that can be integrated with other modalities acquired at the single cell level.
With scPortrait, we
- Mapped dissociated single-cell transcriptomics data to tissues using flow matching
- Identified a morphology-defined subset of immune cells that are associated with ovarian cancer
- Embedded single-cell images across modalities into transcriptome-based atlases
scPortrait Manuscript →
Modeling cell biology experiments with AI
Prophet is a general-purpose AI model for cellular phenotype prediction that generalizes by modeling each cell biology experiment as a composite of three elements:
- A cellular model system the experiment is conducted in
- A perturbation applied to that model system
- A readout that characterises the model systems's response
Image-based genetic screening with SPARCS
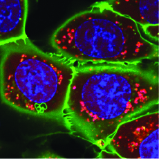
Together with Veit Hornung's and Matthias Mann's lab I developed the SPARCS technology to conduct image-based genetic screens in cell culture models with unprecedented throughput. In SPARCS, we image a library of genetically altered cells on a microscope, generate a single-cell image dataset using scPortrait and then select individual cells with interesting cells from hundreds of millions of single-cell images using AI-based cellular phenotyping methods.
SPARCS enables new types of genetic screens on complex phenotypic readouts such as precise intracellular organelle positions in which selective pressure is defined and applied by AI models. This enables the discovery of entirely new biological mechanisms.
SPARCS Manuscript →